Studying COVID-19 with Air-Liquid Interface Cultures
Why Is ALI a Physiologically Relevant Model System for Viral Infection?
The human airway epithelium constitutes the first line of defense against respiratory pathogens, air pollution, and allergens. Studying how these environmental insults affect the airway tissue and developing effective treatments to combat them requires a physiologically relevant model system.
Conventional submerged culture systems do not adequately capture the complex morphological and functional characteristics of the in vivo human airway, and animal primary cell models can have limited experimental windows and lack perfect homology to human systems. Addressing these limitations, air-liquid interface (ALI) culture of human primary airway epithelial cells is being increasingly adopted as a model system for the in vivo human airway epithelium.
For the study of respiratory viruses, such as the novel SARS-CoV-2 virus that causes COVID-19, ALI cultures are advantageous because they preserve key characteristics of the in vivo airway epithelium that viruses target. Viruses evolve in parallel to their host cells and, consequently, show pronounced specificity for both host species and cell type. Infection models for a given virus must therefore be specific to and closely representative of the native cells it targets. To date, the ALI culture system has been used to study a wide range of viruses, including the novel coronavirus.
Early Studies of the Novel Coronavirus SARS-CoV-2 with ALI Culture
Zhu N et al. (2020) A novel coronavirus from patients with pneumonia in China. N Engl J Med 382(8):727–33.
Lu R et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224): 565–74.
In order to isolate and identify a novel pathogen causing illness in China, researchers involved in the two studies above infected ALI cultures of pathogen-free human airway epithelial cells with fluid from patients showing symptoms. This human airway infection model allowed the scientists to isolate the virus that was reproducibly infecting human lung cells, sequence its genome, and investigate elements of its pathogenesis with transmission electron microscopy.
Zhu N et al. (2020) Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun 11(1): 3910.
Scientists from the Chinese Center for Disease Control and Prevention infected primary human airway epithelial cells (HAECs) with SARS-CoV-2 to investigate transmission, morphogenesis, and pathogenesis of the virus. A pseudostratified epithelial layer, resembling that of the human airway and formed by expanding and differentiating HAECs in PneumaCult™-Ex Plus Medium and PneumaCult™-ALI Medium, respectively, was used to study the virus’ replication efficacy and the cytopathic and morphological changes in ciliated and secretory cells.
- See recent news related to these publications >
Lukassen S et al. (2020) SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J Apr 4. Epub ahead of print, DOI: 10.15252/embj.20105114.
Scientists in Germany used single-cell RNA sequencing (scRNAseq) to probe which cells in the lungs had the highest expression of transmembrane receptors for the SARS-CoV-2 virus. Although they used primary healthy lung tissue samples for the larger airways, this is impractical for the very small subsegmental bronchial branches, where sample sizes are small. To increase the number of cells for analysis, these scientists grew cells from the lung’s smallest airways into ALI cultures in PneumaCult™-ALI Medium.
Sungnak et al. (2020) SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26: 681–87.
In this study, an international group of researchers used scRNAseq and ALI cultures of nasal epithelial cells grown in PneumaCult™-ALI Medium to show that differentiating cells at the ALI, particularly secretory and ciliated cells, express progressively more ACE2. They note the value of the ALI culture system as a biologically relevant in vitro model for SARS-CoV-2 pathogenesis.
Webinar: Studying Respiratory Viruses with ALI Cultures
ALI as a Model System to Study Viruses, Including SARS-CoV-2
ALI culture of primary human airway cells is uniquely suited as a model for in vitro infection studies. This is due to the ability of ALI cultures to faithfully recapitulate key characteristics of the in vivo airway. For example, ALI cultures exhibit relevant proportions of airway cell types, appropriate cellular polarization and junctional properties, dynamic physiologic processes such as mucus secretion and coordinated ciliary beating, and physiological expression and subcellular localization of characteristic proteins bearing species-specific sequences. Depending on the infection mechanism of the virus under study, each one of these properties can be a crucial feature of the model system.
The ability of ALI cultures to model the key features of the in vivo airway highlights the greater physiological relevance of this system over other conventional cell culture models. Human airway ALI cultures allow for efficient infection of cultured cells and have been used to model various mechanisms of viral pathogenesis and human disease (see publication highlights below). This model provides a platform for researchers to study how specific viral characteristics influence pathogenesis during infection. This includes investigating protein interactions during intracellular replication,1 factors impacting viral entry into cells,2 and infection kinetics.3 The ALI infection model has also been used to interrogate the mechanisms of tissue damage and cell death4 as well as aspects of host response, such as interferon response to infection.5 The applications of ALI cultures, which can be generated from pediatric6 and adult human cells, are wide ranging. ALI cultures have been co-infected with bacteria and viruses, and cultures generated from zoonotic vectors (e.g. pig) have also been studied in the context of human disease dynamics.7 The utility of ALI cultures for studying viral infection represents a valuable tool for developing antiviral therapeutic approaches;5 viral neutralization by monoclonal antibodies has also been tested in this model.8
Coronaviruses (CoV) are a large family of viruses that cause illnesses ranging from the common cold to severe respiratory diseases, including COVID-19 caused by the SARS-CoV-2 virus. The cell entry receptor for SARS-CoV-2 has recently been confirmed by independent research groups to be angiotensin-converting enzyme 2 (ACE2)—the same entry receptor for the SARS-CoV virus that caused the 2003 SARS outbreak9,10. Human large and small airway cultures grown at the ALI show abundant ACE2 expression (Figure 1) and provide a physiologically relevant model for investigating ACE2 as a target for antiviral intervention against COVID-19.
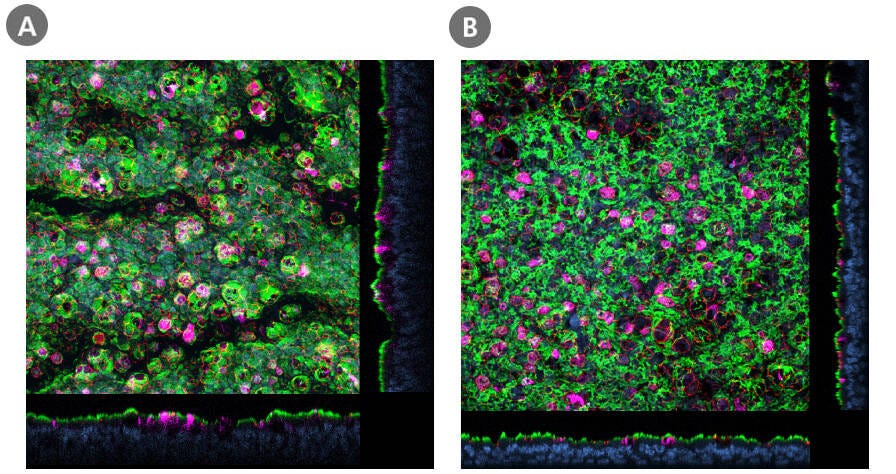
Figure 1. Presence of ACE2 in HBECs Cultured in PneumaCult™-ALI Medium and HSAECs Cultured in PneumaCult™-ALI-S Medium
Confocal images of whole mount immunostained ALI cultures of (A) human bronchial epithelial cells (HBECs) cultured in PneumaCult™-ALI Medium and (B) human small airway epithelial cells (HSAECs) cultured in PneumaCult™-ALI-S Medium, at P2. The ALI cultures were fixed and stained with antibodies for ciliated cells (AC-tubulin; green), SARS-CoV-2 receptor (ACE2; magenta), and tight junctions (ZO1; red). The nuclei were counterstained with DAPI (blue). ACE2 was detected in both (A) large and (B) small airway culture systems during early passages (≤P6, HBECs; ≤P4, HSAECs). Both HBECs and HSAECs were expanded in PneumaCult™-Ex Plus Medium prior to differentiation at the ALI.
Publication Highlights
Coronavirus (CoVs)
- Menachery VD et al. (2020) Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J Virol 94(5): e01774–19.
- Widagdo W et al. (2019) Species-specific colocalization of Middle East respiratory syndrome coronavirus attachment and entry receptors. J Virol 93(16): e00107–19.
- Shirato K et al. (2018) Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 517: 9–15.
Parvovirus (HBoV1)
- Wang K et al. (2020) Cellular cleavage and polyadenylation specificity factor 6 (CPSF6) mediates nuclear import of human bocavirus 1 NP1 protein and modulates viral capsid protein expression. J Virol 94(2): e01444–19.
- Deng et al. (2017) Human parvovirus infection of human airway epithelia induces pyroptotic cell death by inhibiting apoptosis. J Virol 91(24):e01533–17.
Respiratory syncytial virus (RSV)
- Wethington D et al. (2019) Mathematical modelling identifies the role of adaptive immunity as a key controller of respiratory syncytial virus in cotton rats. J R Soc Interface. 16(160):20190389.
- Anderson CS et al. (2019) CX3CR1 as a respiratory syncytial virus receptor in pediatric human lung. Pediatr Res doi: 10.1038/s41390-019-0677-0.
- Cortjens B et al. (2017) Broadly reactive anti-respiratory syncytial virus G antibodies from exposed individuals effectively inhibit infection of primary airway epithelial cells. J Virol 91(10): e02357–16.
Influenza
- Richard M et al. (2020) Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat Commun 7;11(1):766.
- Chen AJ et al. (2019) Anti-H7N9 avian influenza A virus activity of interferon in pseudostratified human airway epithelium cell cultures. J Virol 16(1):44.
- Meng F et al. (2019) Viral coinfection replaces effects of suilysin on Streptococcus suis adherence to and invasion of respiratory epithelial cells grown under air-liquid interface conditions. Infect Immun 87(8): e00350–19.
Rhinovirus
- Wu Q et al. (2018) Overproduction of growth differentiation factor 15 promotes human rhinovirus infection and virus-induced inflammation in the lung. Am J Physiol Lung Cell Mol Physiol 314(3): L514–L527.
- Griggs TF et al. (2017) Rhinovirus C targets ciliated airway epithelial cells. Respir Res 18(1): 84.
- Warner SM et al. (2019) Rhinovirus replication and innate immunity in highly differentiated human airway epithelial cells. Respir Res 20(1):150.
Why Is ALI Culture Physiologically Relevant?
Primary human bronchial epithelial cells (HBECs) cultured at the ALI undergo extensive mucociliary differentiation that recapitulates what is observed in vivo. As a result, ALI culture of HBECs is increasingly being recognized as the most physiologically relevant in vitro model system for respiratory research. While the use of immortalized cell lines and primary cells from animals is common, data generated using these models are not directly applicable to the human system.11 Submerged culture of primary HBECs is possible; however, cells in this system fail to undergo mucociliary differentiation and thus do not accurately represent the in vivo airway.
The physiological relevance of the ALI culture system is supported by phenotypical and histological evidence. Hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS) staining show that HBECs cultured at the ALI are (Figure 2A and 2C) pseudostratified in morphology and make up a heterogeneous cell population that includes ciliated and mucus-secreting (PAS-positive) cells, similar to the in vivo bronchial epithelium (Figure 2B and 2D). An important function of the tracheobronchial epithelium in vivo is to act as a protective barrier against inhaled insults. The same epithelial barrier function has been confirmed in ALI cultures by the expression of tight junction proteins and the development of high transepithelial electrical resistance.12 The suitability of ALI cultures for modeling the airway has been further confirmed through transcriptome analyses13 and a wide range of experiments demonstrating physiological responses to insults such as toxicants and pathogens.14-16 Importantly, ALI cultures of primary cells from donors with respiratory disease (e.g. asthma, cystic fibrosis, COPD) recapitulate in vivo disease characteristics, forming robust in vitro models of these conditions.17-18
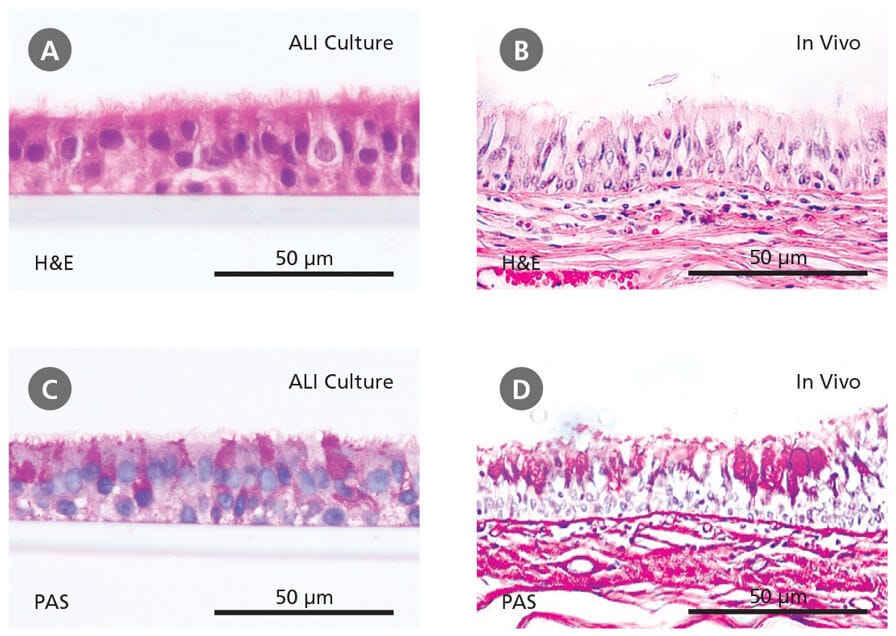
Figure 2. Primary Human Bronchial Epithelial Cells Cultured at the Air-Liquid Interface Recapitulate the In Vivo Bronchial Epithelium
Hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS) staining reveals that cells differentiated at the air-liquid interface in PneumaCult™-ALI (A and C) form a pseudostratified epithelium that is representative of the in vivo bronchial epithelium (B and D). Data in (A) and (C) generated by Samuel Wadsworth.
Webinar: In Vitro Human Airway Modeling Using Primary Airway Epithelial Cells
How Are Cells Grown at the ALI?
The defining feature of ALI culture is that the basal surface of the cells is in contact with liquid culture medium, whereas the apical surface is exposed to air. A common approach is to seed cells onto the permeable membrane of a cell culture insert, which is initially supplied with culture medium to both the apical and basal compartments (Figure 3A). Once confluence is reached, the cells are subjected to ‘air-lift’, where the medium is supplied only to the basal chamber (Figure 3B). This configuration mimics the conditions found in the human airway and drives differentiation towards a mucociliary phenotype.
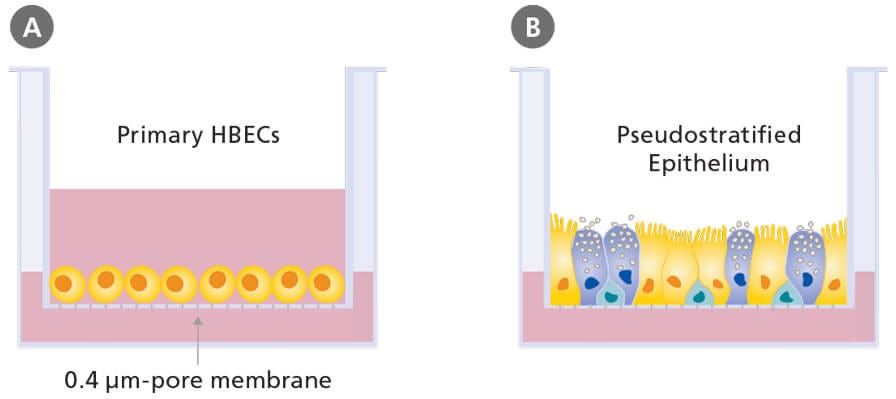
Figure 3. Air-Liquid Interface Culture
Schematic of submerged (A) and air-liquid interface (B) culture of primary human bronchial epithelial cells grown using porous culture inserts.
Products for Pulmonary Research
Recombinant Spike, Nucleocapsid, and Human ACE2 Protein
Products for Targeting SARS-CoV-2 Nucleoprotein and Spike Protein
Explore more tools, products and support for COVID-19 and SARS-CoV-2 research.
Featured Content and Resources
Respiratory System Overview
Get a free wallchart providing an organizational and functional overview of the respiratory system.
Optimizing In Vitro Lung Models
Dr. Rachael Rayner compares the effects of various expansion and differentiation media on primary airway cells cultured at the ALI.
References
- Wang K et al. (2020) Cellular cleavage and polyadenylation specificity factor 6 (CPSF6) mediates nuclear import of human bocavirus 1 NP1 protein and modulates viral capsid protein expression. J Virol 94(2): e01444–19.
- Griggs TF et al. (2017) Rhinovirus C targets ciliated airway epithelial cells. Respir Res 18(1): 84.
- Wethington D et al. (2019) Mathematical modelling identifies the role of adaptive immunity as a key controller of respiratory syncytial virus in cotton rats. J R Soc Interface. 16(160):20190389.
- Deng et al. (2017) Human parvovirus infection of human airway epithelia induces pyroptotic cell death by inhibiting apoptosis. J Virol 91(24):e01533–17.
- Chen AJ et al. (2019) Anti-H7N9 avian influenza A virus activity of interferon in pseudostratified human airway epithelium cell cultures. J Virol 91(24)
- Anderson CS et al. (2019) CX3CR1 as a respiratory syncytial virus receptor in pediatric human lung. Pediatr Res Nov 14.
- Meng F et al. (2019) Viral coinfection replaces effects of suilysin on Streptococcus suis adherence to and invasion of respiratory epithelial cells grown under air-liquid interface conditions. Infect Immun 87(8): e00350–19.
- Cortjens B et al. (2017) Broadly reactive anti-respiratory syncytial virus G antibodies from exposed individuals effectively inhibit infection of primary airway epithelial cells. J Virol 91(10): e02357–16.
- Zhou et al. (2020) A Pneumonia Outbreak Associated With a New Coronavirus of Probable Bat Origin. Nature 579 (7798): 270-273.
- Hoffmann et al. (2020) SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell March 4.
- Bérubé K et al. (2010) Human primary bronchial lung cell constructs: the new respiratory models. Toxicology 278(3): 311-318
- Prytherch Z et al. (2011)Tissue-Specific stem cell differentiation in an in vitro airway model. Macromol Biosci 11(11): 1467-1477
- Dvorak A et al. (2011) Do airway epithelium air-liquid cultures represent the in vivo airway epithelium transcriptome? Am J Respir Cell Mol Biol 44(4): 465-473
- Thaikoottathil JV et al. (2009) Cigarette smoke extract reduces VEGF in primary human airway epithelial cells. Eur Respir J 33(4): 835-843
- Krunkosky TM et al. (2007)Mycoplasma pneumoniae host-pathogen studies in an air-liquid culture of differentiated human airway epithelial cells. Microb Pathog 42(2-3): 98-103
- Palermo LM et al. (2009)Human parainfluenza virus infection of the airway epithelium: viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J Virol 83(13): 6900-6908
- Ostrowski LE et al. (2012) Interferon γ stimulates accumulation of gas phase nitric oxide in differentiated cultures of normal and cystic fibrosis airway epithelial cells. Lung 190(5): 563-571
- Comer D et al. (2013) Airway epithelial cell apoptosis and inflammation in COPD, smokers and nonsmokers. Eur Respir J doi: 10.1183/09031936.00063112
- Yan et al. (2017) Human Bocavirus Type-1 Capsid Facilitates the Transduction of Ferret Airways by Adeno-Associated Virus Genomes. Hum Gene Ther. Aug;28(8):612-625.
- Tata et al.(2013) Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. Nov 14;503(7475):218-23.
- Dale et al. (2019) Rock Inhibition Y-27632 Enables Feeder-Free, Unlimited Expansion of Sus scrofa domesticus Swine Airway Stem Cells To Facilitate Respiratory Research. Nov 23;2019:3010656.
- Engler et al.(2019) Isolation, Maintenance and Differentiation of Primary Tracheal Basal Cells from Adult Rhesus Macaque. Methods Protoc. Oct 1;2(4). pii: E79.