CRISPR-Cas9 Genome Editing of Human NK Cells
Natural Killer (NK) cells are innate immune cells capable of killing tumor or virus-infected cells as well as secreting pro-inflammatory cytokines. While genome editing of NK cells remains challenging, this protocol highlights an ex vivo method to overcome some of these challenges. CRISPR-Cas9, an RNA-guided genome editing technology, is revolutionizing cell biology due to the ease and efficiency by which it enables the genetic manipulation of mammalian cells. Through targeted modifications of specific genes or regulatory regions, researchers can now rapidly generate precise genetic models to study normal and diseased cell physiology. Beyond genetic manipulation for research purposes, CRISPR-Cas9 genome editing also holds great potential for therapeutic applications, including immunotherapy and regenerative medicine.
Below, we describe a protocol for CRISPR-Cas9 genome editing of primary human NK cells cultured in ImmunoCult™ NK Cell Expansion Kit (Catalog #100-0711) using the ArciTect™ CRISPR-Cas9 ribonucleoprotein (RNP)-based system and STEMCELL’s Guide RNA Design Tool. The protocol also details instructions for the isolation and activation of primary human NK cells, preparation of a CRISPR-Cas9 RNP complex, and delivery of an RNP complex into activated primary NK cells via electroporation using either the Neon® Transfection System or Lonza® 4D Nucleofector™ X Unit.
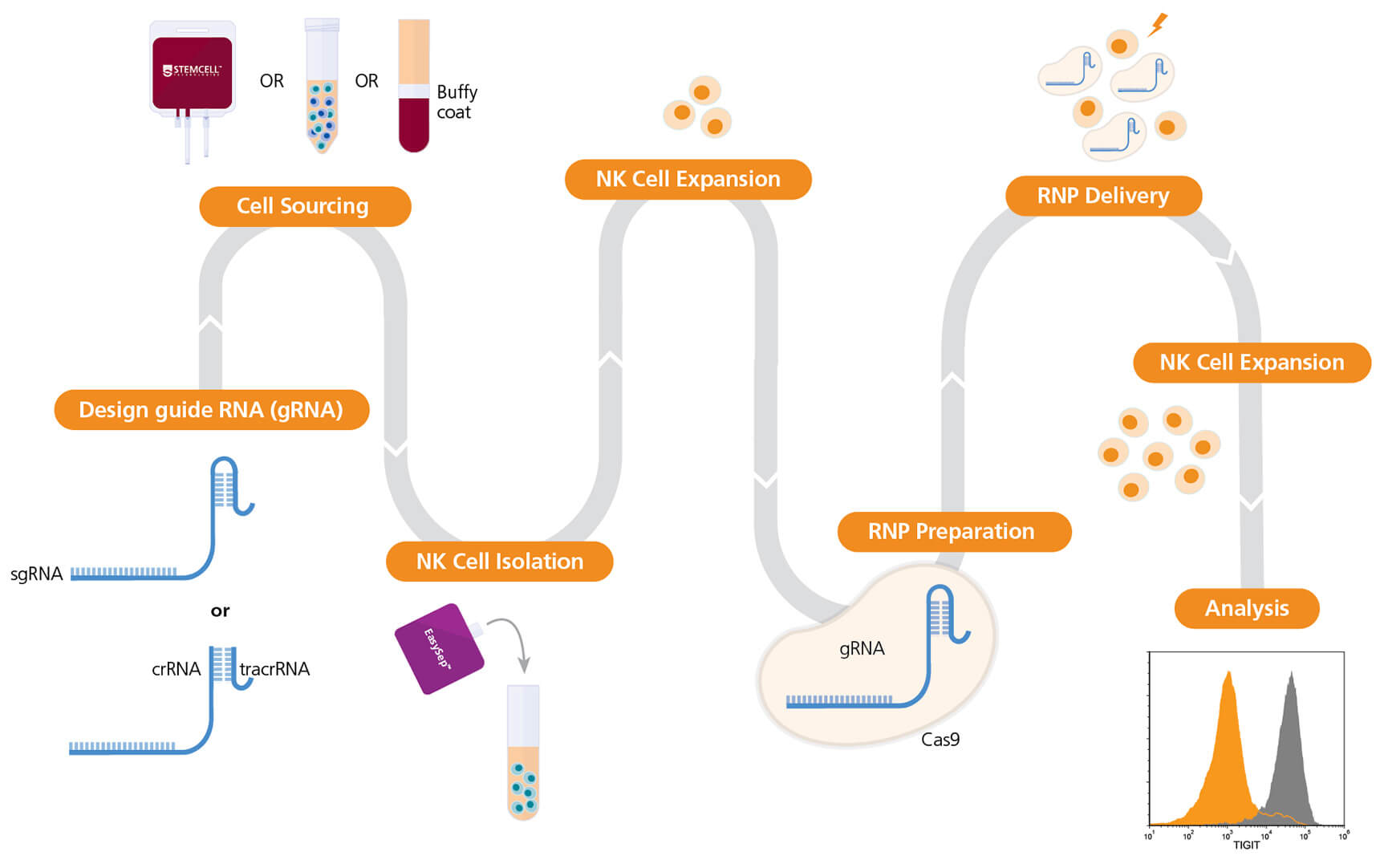
Figure 1. Experimental Workflow for Human Natural Killer (NK) Cell Genome Editing
The ArciTect™ sgRNA (single guide RNA) or ArciTect™ crRNA (CRISPR RNA) sequences can be designed using the CRISPR Design Tool once a target locus for editing is identified. NK cells can be purchased or isolated from a number of sources, such as cord blood, bone marrow, and mobilized peripheral blood, using column-free cell separation technology, including EasySep™. Next, NK cells are cultured for 3-4 days on a plate pre-coated with ImmunoCult™ NK Cell Expansion Coating Material (1X) prior to ribonucleoprotein (RNP) delivery in Complete ImmunoCult™ NK Cell Expansion Medium. The ArciTect™ CRISPR-Cas9 RNP is then prepared and delivered into NK cells using electroporation, and cells are plated in Complete ImmunoCult™ NK Cell Expansion Medium post-electroporation. Editing efficiency can be analyzed using flow cytometry.
Materials
- ArciTect™ Cas9 Nuclease (Catalog #76002/76004)
- ArciTect™ sgRNA† (Catalog #200-0013)
- Nuclease-Free Water (Catalog #79001)
- EasySep™ Human NK Cell Isolation Kit (Catalog #17955)
- ImmunoCult™ NK Cell Expansion Kit (Catalog #100-0711)
- ImmunoCult™ NK Cell Base Medium (Catalog #100-0712)
- ImmunoCult™ NK Cell Expansion Supplement (100X) (Catalog #100-0715)
- ImmunoCult™ NK Cell Expansion Coating Material (100X) (Catalog #100-0714)
- D-PBS (Without Ca++ and Mg++) (Catalog #37350)
- Neon® Transfection System 10 μL Kit (Thermo Fisher MPK1025)
- Resuspension Buffer R
- Electrolytic Buffer E
- 10 μL Neon® Pipette Tips OR
- P3 Primary Cell 4D-Nucleofector™ X Kit S (Lonza® V4XP-3032)
- 16-Well Nucleocuvette™ Strips
- P3 Primary Cell Nucleofector™ Solution
- Supplement 1
Part I: Preparation of Complete ImmunoCult™ NK Cell Expansion Medium
Use sterile technique to prepare Complete ImmunoCult™ NK Cell Expansion Medium (ImmunoCult™ NK Cell Base Medium + ImmunoCult™ NK Cell Expansion Supplement [100X]). The following example is for preparing 10 mL of complete medium.
- Thaw ImmunoCult™ NK Cell Expansion Supplement (100X) at room temperature (15 - 25°C) or overnight at 2 - 8°C. Mix thoroughly.
Note: If not used immediately, aliquot and store ImmunoCult™ NK Cell Expansion Supplement at -20°C. Alternatively, store at 2 - 8°C for up to 1 month. Do not exceed the shelf life of the supplement. After thawing the aliquots, use immediately. Do not re-freeze.
- Add 100 μL of ImmunoCult™ NK Cell Expansion Supplement (100X) to 9.9 mL of ImmunoCult™ NK Cell Base Medium. Mix thoroughly.
Note: If not used immediately, store complete ImmunoCult™ NK Cell Expansion Medium at 2 - 8°C for up to 2 weeks.
Part II: Preparation of ImmunoCult™ NK Cell Expansion Coating Material (1X)
Use sterile technique to prepare ImmunoCult™ NK Cell Expansion Coating Material (1X) (ImmunoCult™ NK Cell Expansion Coating Material [100X] + D-PBS [Without Ca++ and Mg++]). The following example is for preparing 10 mL of coating material. If preparing other volumes, adjust accordingly.
Add 100 μL of ImmunoCult™ NK Cell Expansion Coating Material (100X) to 9.9 mL of sterile D-PBS. Mix thoroughly. Use immediately.
Part III: NK Cell Isolation and Activation
- Add 500 μL of ImmunoCult™ NK Cell Expansion Coating Material (1X) per well of a non-tissue culture-treated 24-well plate.
- Incubate at room temperature (15 - 25°C) for 2 hours.
Note: Alternatively, coating can be conducted overnight in the fridge.
- Aspirate coating material from the 24-well plate. Rinse the well with D-PBS. Aspirate D-PBS just prior to use.
- Isolate human NK cells from peripheral blood using the EasySep™ Human NK Cell Isolation Kit (Catalog #17955).
- Perform a viable cell count using Trypan Blue and a hemocytometer.
- Add NK cells at 1 x 10⁶ cells/mL to 500 μL of complete ImmunoCult™ NK Cell Expansion Medium.
- Add 500 μL of cell suspension (prepared in step 6) (i.e. 5 x 10⁵ cells) to one coated well of the 24-well plate prepared in step 3. Incubate at 37°C and 5% CO₂ for 3 or 4 days.
- Carefully add 500 μL of complete ImmunoCult™ NK Cell Expansion Medium per well of the 24-well plate. Incubate at 37°C and 5% CO₂ for 3 to 4 days.
Part IV: Preparation of ArciTect™ sgRNA Working Solution
- Briefly centrifuge the ArciTect™ sgRNA vials before opening.
- Add nuclease-free water to each vial to give a final concentration of 100 μM single guide RNA (sgRNA), as indicated in Table 1.
Table 1. Resuspension Volume for 100 μM* ArciTect™ sgRNA
AMOUNT OF ArciTect™ sgRNA (nmol)VOLUME OF NUCLEASE-FREE WATER (µL)440*100 µM is equal to 100 pmol/µL
- Mix thoroughly. If not used immediately, aliquot and store at -20°C for up to 6 months or at -80°C for longer than 6 months. After thawing the aliquots, use immediately. Do not refreeze.
Part V: Preparation of ArciTect™ CRISPR-Cas9 RNP Complex Mix for Electroporation
- To prepare the RNP Complex Mix, combine the components, in the order listed in Table 2, in a microcentrifuge tube. Adjust component volumes according to the desired number of transfections.
- Mix thoroughly.
Table 2. Preparation of RNP Complex Mix for Electroporation
Component sgRNA Neon® Electroporation 4D-Nucleofector™ X Electroporation Volume per Reaction (μL) Resuspension Buffer R 6.18 -- P3 Primary CellNucleofector™ Solution with Supplement 1 -- 5.3 ArciTect™ Cas9 Nuclease (4 μg/μL; 25 μM) 0.96 1.6 100 μM sgRNA 0.36 0.60 Total 7.50 7.50 Note: May require optimization with different cell sources. A 1:2.67 (shown) to 1:8 molar ratio of Cas9 to guide RNA is recommended.Note: These values are for a single electroporation reaction and include pipetting error for Neon® electroporation. Scale up as necessary. - Incubate the RNP Complex Mix at room temperature (15 - 25°C) for 20 minutes.
Part VI: Electroporation of Activated NK cells with RNP complex
Perform electroporation using either the Neon® Transfection System (section a) or the Lonza® 4D-Nucleofector™ X Unit (section b).
a) Electroporation Using Neon® Transfection System
- Harvest NK cells (from part III), count cells, and transfer 300,000 cells to a 15 mL Falcon tube.
- Centrifuge at 300 x g for 5 mins. Aspirate supernatant from the cell pellet. Resuspend 300,000 cells in 7.5 μL of Resuspension Buffer R per electroporation parameter and pipette up and down vigorously to mix.
- Transfer 7.5 μL of the cell suspension to each 7.5 μL of RNP Complex Mix (prepared in part IV) and pipette up and down gently to mix, trying not to form air bubbles.
- Using a 10 μL Neon® pipette tip, draw up 10 μL of the mixture, ensuring the capillary is free of bubbles, and place it into the electroporation chamber containing 3 mL of Electrolytic Buffer E.
Note: If air bubbles are present in the tip when the cells are electroporated, cell viability and transfection efficiency will be significantly reduced.
- Electroporate the mixture using the settings in Table 3.
Note: Refer to the manufacturer’s instructions on electroporation. Electroporation parameters may require optimization for different cell sources.
Table 3. Recommended Electroporation Parameters for NK Cells Using a Neon® Transfection System
Electroporation Parameters Electrical potential 1800 V Pulse width 20 ms Number of pulses 1
b) Electroporation Using Lonza® 4D-Nucleofector™ X Unit
- Harvest NK cells (from part III), count cells, and transfer 500,000 cells to a 15 mL Falcon tube.
- Centrifuge at 300 x g for 5 mins. Aspirate supernatant from the cell pellet. Resuspend 500,000 cells in 17.5 μL of P3 Primary Cell Nucleofector™ Solution + Supplement 1 per electroporation parameter and pipette up and down vigorously to mix.
- Transfer 17.5 μL of the cell suspension to each 7.5 μL RNP Complex Mix (prepared in part IV) and pipette up and down gently to mix, trying not to form air bubbles.
Note: If air bubbles are present in the cuvette when the cells are electroporated, cell viability and transfection efficiency will be significantly reduced.
- Transfer 25 μL of the cell suspension + RNP Complex Mix to one well of the 16-well Nucleocuvette™ Strip. Gently tap or use a pipette tip to ensure no air bubbles are present.
- Set the Lonza® 4D-Nucleofector™ X Unit to program code CM-137.
- Place the Nucleocuvette™ Strip in the Shuttle device of the 4D-Nucleofector™ X Unit, select OK to load the strip, and select Start to begin electroporation.
Part VII: Post-Editing Culture
- Immediately add the electroporated cells to 1 well of the 24-well plate prepared in part V.
- Incubate at 37°C and 5% CO₂ for 4 - 7 days for genome editing to occur. Harvest cells for assessment of genome editing efficiency.
Note: For further expansion and to reduce the risk of overgrowth and cell death, feed and split the cells at 1 million cells/mL.Note: Genomic DNA can be amplified by PCR using primers flanking the target region and ArciTect™ High-Fidelity DNA Polymerase Kit (Catalog #76026), followed by sequencing of the PCR products. Alternatively, ArciTect™ T7 Endonuclease I Kit (Catalog #76021) can be used to assess editing efficiency (% INDEL formation) following PCR amplification.
Data
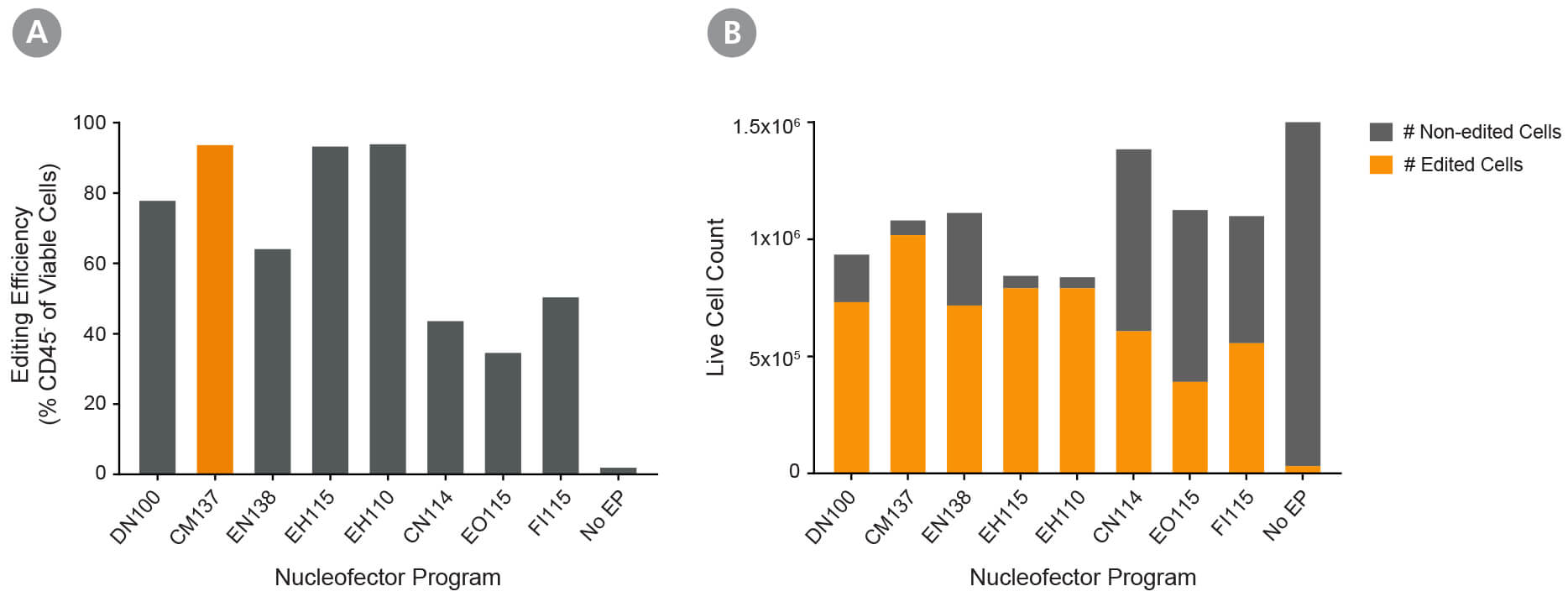
Figure 2. Optimization of Fresh Primary Activated NK Cell RNP Delivery Using the 4D- Nucleofector™ System
Fresh primary NK cells were expanded using ImmunoCult™ NK Cell Expansion Kit. RNP complexes containing ArciTect™ Cas9 Nuclease and ArciTect™ sgRNA targeting the CD45 gene were delivered using the indicated 4D-Nucleofector™ program, and editing efficiency was monitored by flow cytometry to detect surface expression of CD45. (A) Editing efficiency was measured 4 days after electroporation by flow cytometry using a fluorophore-conjugated CD45 antibody to detect functional knockout of CD45. (B) The total number of edited cells (orange bars) and non-edited NK cells (grey bars) was calculated by multiplying editing efficiency by post-electroporation recovery. This indicated which electroporation programs resulted in the best overall editing efficiency and cell recovery. Based on this data, CM137 was selected as the optimal 4D-Nucleofector program.
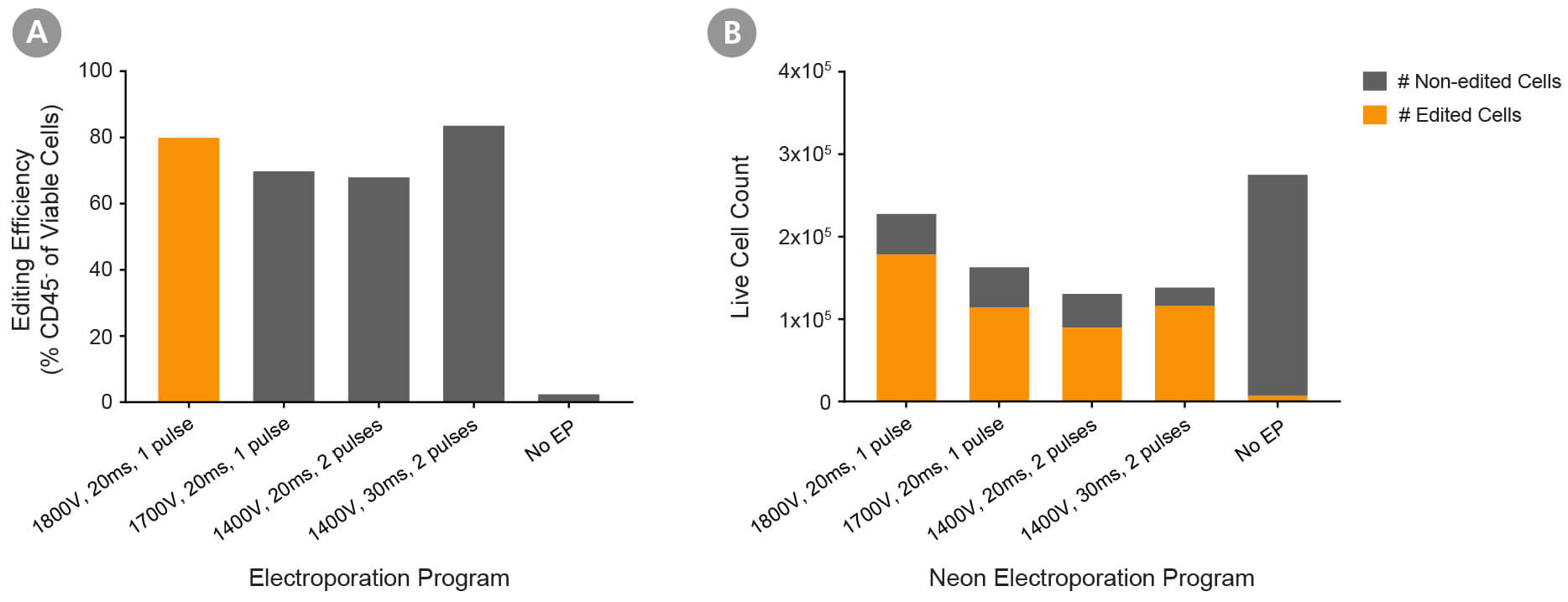
Figure 3. Optimization of Fresh Primary Activated NK Cell RNP Delivery Using the Neon® Transfection System 10 μL Kit
Fresh primary NK cells were expanded using ImmunoCult™ NK Cell Expansion Kit. RNP complexes containing ArciTect™ Cas9 Nuclease and ArciTect™ sgRNA targeting the CD45 gene were delivered using the indicated electroporation programs, and editing efficiency was monitored by flow cytometry to detect surface expression of CD45. (A) Editing efficiency was measured 4 days after electroporation by flow cytometry using a fluorophore-conjugated CD45 antibody to detect functional knockout of CD45. (B) The total number of edited cells (orange bars) and non-edited NK cells (grey bars) was calculated by multiplying editing efficiency by post-electroporation recovery. This indicated which electroporation programs resulted in the best overall editing efficiency and cell recovery. Based on this data, 1800 V, 20 ms, and 1 pulse were selected as the recommended electroporation parameters for NK cells using a Neon® Transfection System.
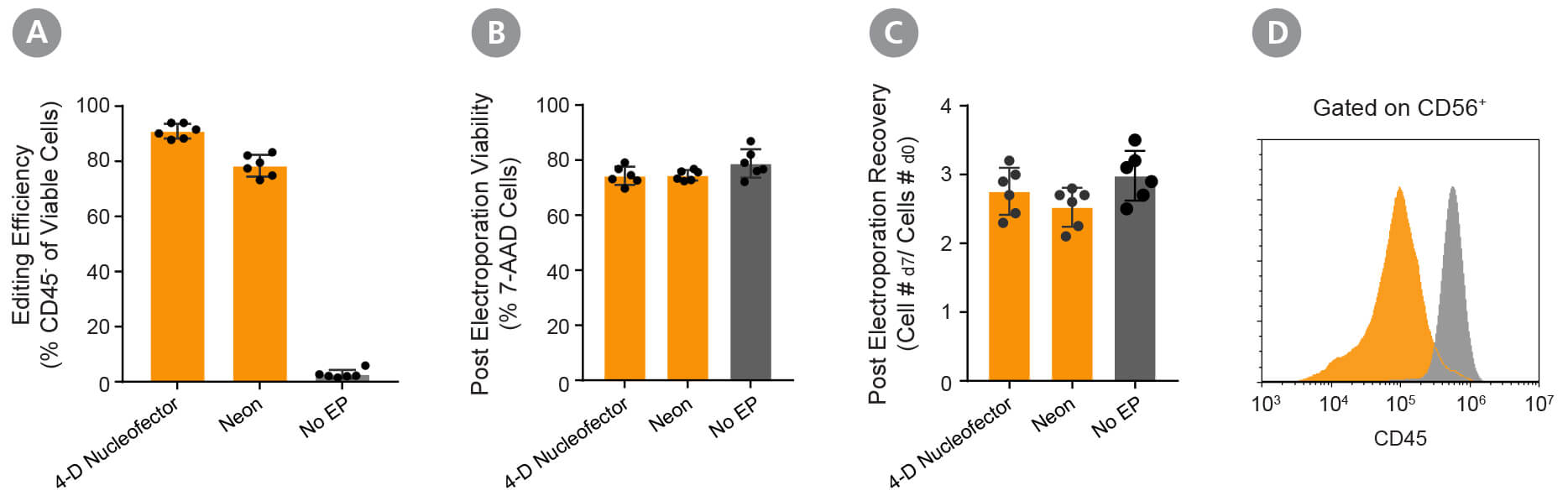
Figure 4. Validation of the Optimized RNP Delivery Electroporation Programs for Activated NK Cells Using ArciTect™ Cas9 Nuclease and ArciTect™ sgRNA Targeting CD45.
Fresh primary NK cells were expanded using ImmunoCult™ NK Cell Expansion Kit. RNP complexes containing ArciTect™ Cas9 Nuclease and ArciTect™ sgRNA targeting the CD45 gene were delivered using either the 4D-Nucleofector™ System (program code: CM-137) or the Neon® Transfection System (1800 V, 20 ms, 1 pulse), and editing efficiency was monitored by flow cytometry to detect surface expression of CD45. (A) Editing efficiency was measured 7 days after electroporation by flow cytometry using a fluorophore-conjugated CD45 antibody to detect functional knockout of CD45. (B) Cell viability was assessed 7 days after electroporation by flow cytometry using 7-AAD dye exclusion for electroporated cells (orange) and non-electroporated cells (grey). (C) Post-electroporation recovery was calculated for both electroporated cells (orange) and non-electroporated cells (grey) by dividing the total number of cells collected 7 days after electroporation by the number of electroporated cells. Each data point represents an individual donor; n = 6 donors. (D) Representative histogram of CD45 flow cytometry data from RNP-electroporated cells using the 4D- Nucleofector™ System (RNP, orange; CM-137) and non-electroporated (No EP; grey) activated NK cells 4 days after delivery of the CRISPR-Cas9 RNP complexes containing sgRNA targeting CD45.
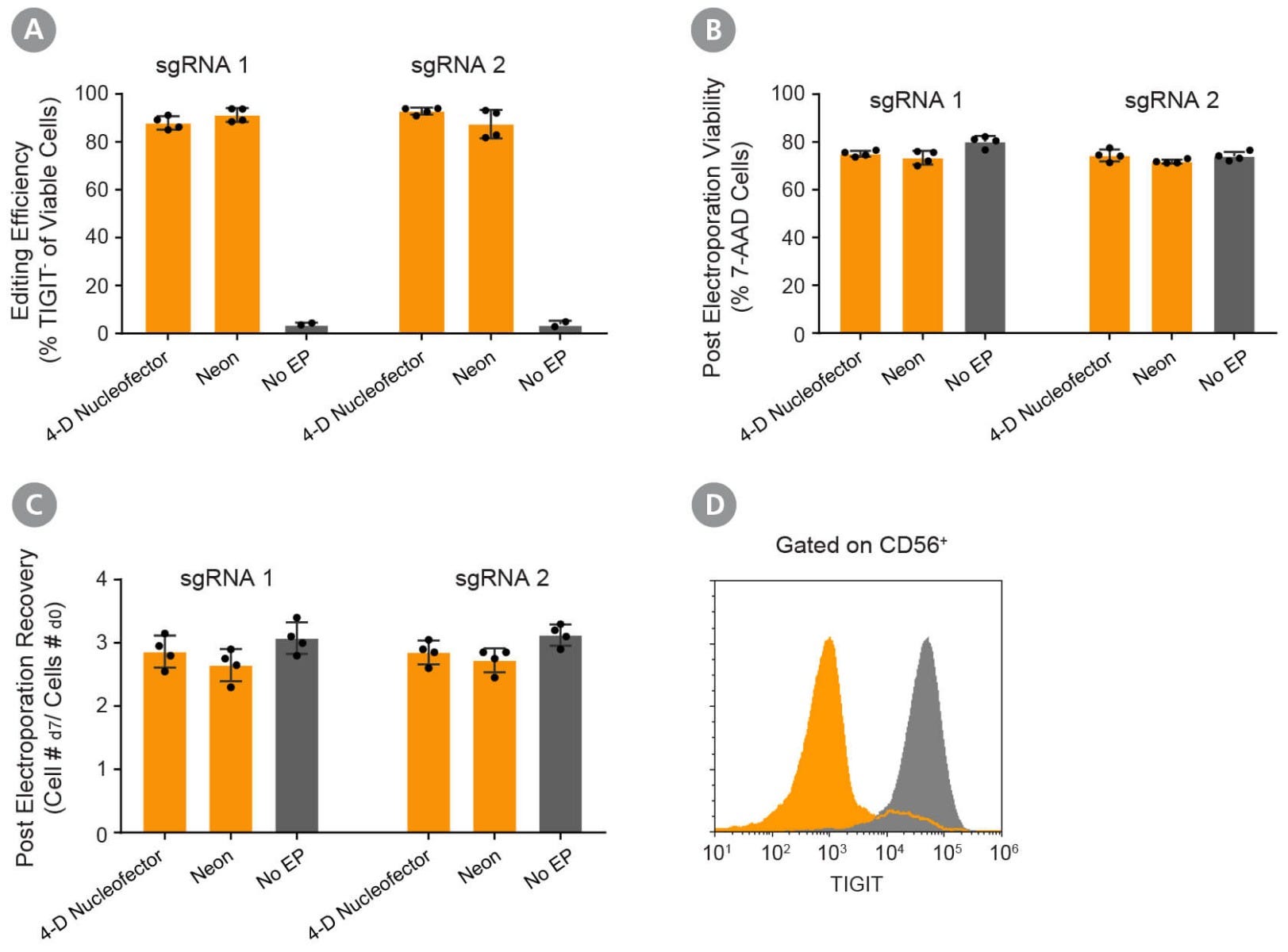
Figure 5. Validation of the Optimized RNP Delivery Electroporation Programs for Activated NK Cells Using ArciTect™ Cas9 Nuclease and ArciTect™ sgRNA Targeting TIGIT at 2 Different Loci.
Fresh primary NK cells were expanded using ImmunoCult™ NK Cell Expansion Kit. RNP complexes containing ArciTect™ Cas9 Nuclease and ArciTect™ sgRNA targeting the TIGIT gene were delivered using either the 4D-Nucleofector™ System (program code: CM-137) or the Neon® Transfection System (1800 V, 20 ms, 1 pulse), and editing efficiency was monitored by flow cytometry to detect surface expression of TIGIT. (A) Editing efficiency was measured 7 days after electroporation by flow cytometry using a fluorophore-conjugated TIGIT antibody to detect functional knockout of TIGIT. (B) Cell viability was assessed 7 days after electroporation by flow cytometry using 7-AAD dye exclusion for electroporated cells (orange) and non-electroporated cells (grey). (C) Post-electroporation recovery was calculated for both electroporated cells (orange) and non-electroporated cells (grey) by dividing the total number of cells collected 7 days after electroporation by the number of electroporated cells. Each data point represents an individual donor; n = 4 donors. (D) Representative histogram of TIGIT flow cytometry data from RNP-electroporated cells using the 4D- Nucleofector™ System (RNP, orange; 1800V, 20 ms, 1pulse) and non-electroporated (No EP; grey) activated NK cells 7 days after delivery of the CRISPR-Cas9 RNP complexes containing sgRNA targeting TIGIT.
Explore These Resources
Request Pricing
Thank you for your interest in this product. Please provide us with your contact information and your local representative will contact you with a customized quote. Where appropriate, they can also assist you with a(n):
Estimated delivery time for your area
Product sample or exclusive offer
In-lab demonstration