Survey Report: Hurdles of Genome Editing Using CRISPR-Cas9
Introduction
Through targeted modifications of specific genes or regulatory regions, researchers can now study gene function, generate disease models, tag genes endogenously, perform gene or cell therapy, and more. Despite the widespread adoption of CRISPR gene editing technology, many challenges remain in successfully applying this tool. Understanding the challenges that researchers are facing in the lab can help the scientific community to collectively find the appropriate solutions.
We conducted a survey of over 300‡ researchers to ask about the issues and concerns they encounter when applying CRISPR genome editing technology in their studies. This report summarizes the survey findings, which showed that researchers are inspired by the promise of CRISPR technology, however, as the applications continue to grow there are specific challenges that need to be addressed to empower researchers to perform high-efficiency genome editing in various cell types including stem and primary cells.
Who participated in the survey?
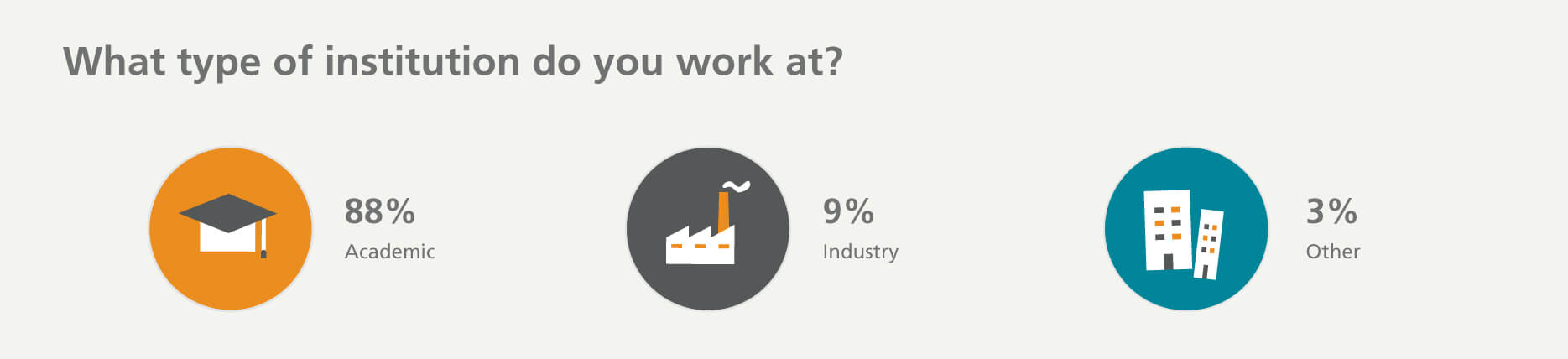
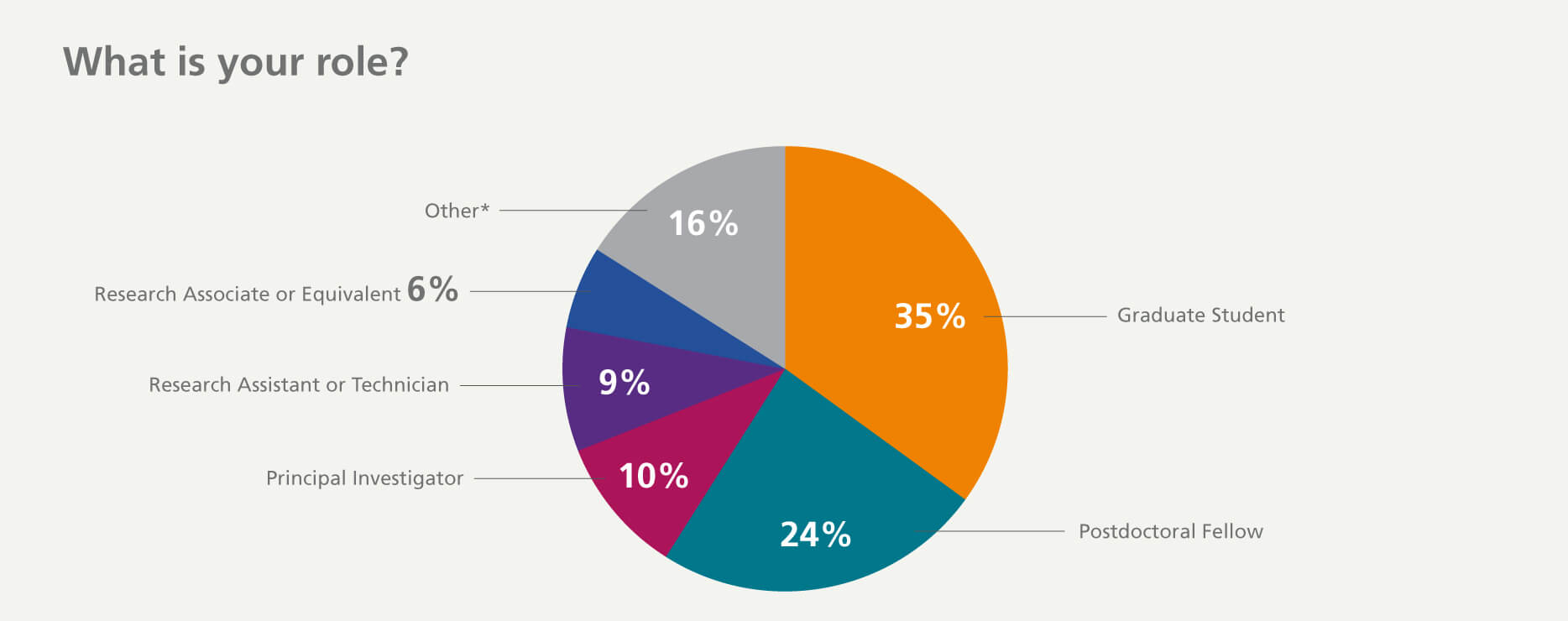
*Others include lab managers, core managers, directors, program managers, CEOs, undergraduate students, interns, QA or QC roles, and procurement and purchasing managers.
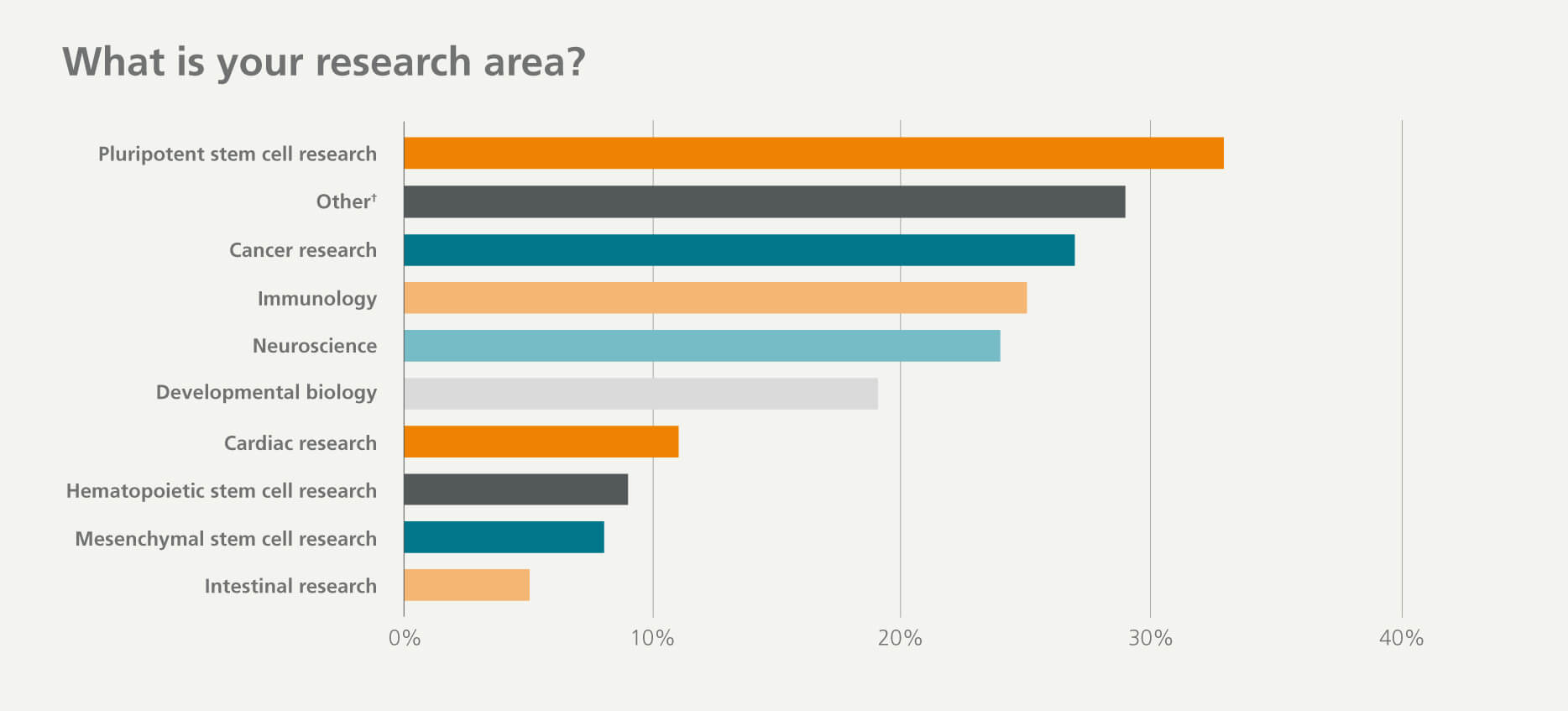
†Others include respiratory research, liver research, mammary cell research, myogenic stem and progenitor cell research, pancreatic research, and endothelial and angiogenic cell research.
‡The survey results were collected in early 2019 and had a total of 345 respondents.
The Promise of CRISPR-Cas9
CRISPR-Cas9 has been hailed as a revolutionary genome editing tool with a wide array of applications, including enabling unprecedented precision in studying gene function and developing improved cell culture models and animal models of disease. It is also rapidly being incorporated into the development of next-generation cellular therapies.
In our survey of life science researchers, 93% of respondents agree that CRISPR-Cas9 technology can benefit their research.
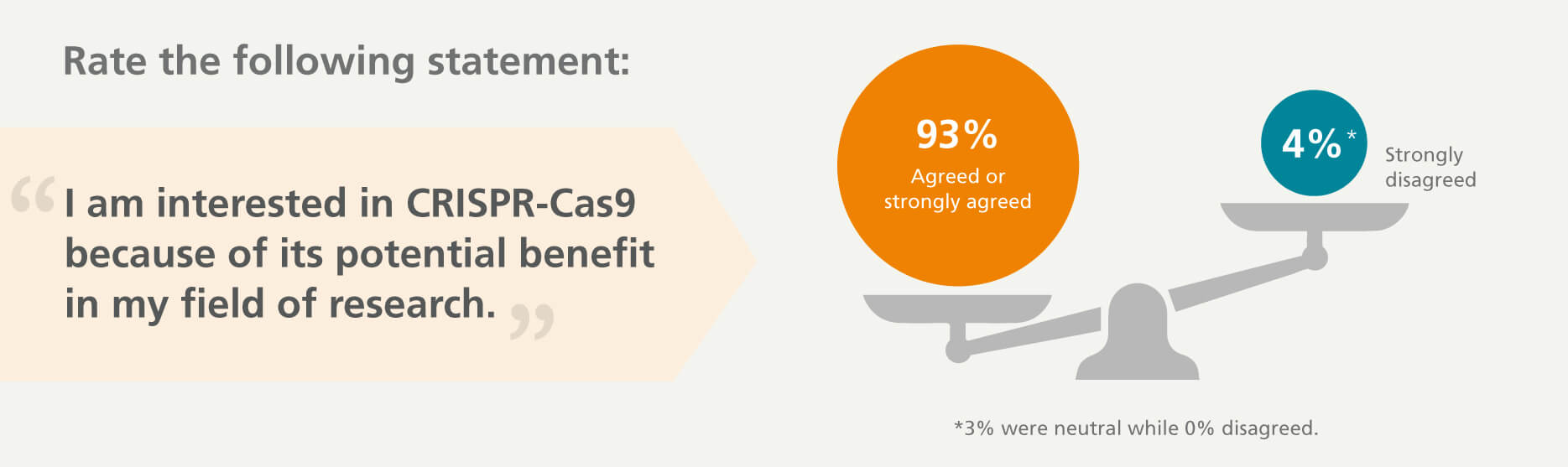
Genome editing will revolutionize medicine — and the world.
Survey Respondent
CRISPR-Cas9 Is Widely Used
Compared to genetic tools such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), CRISPR-Cas9 has greatly improved the ease and efficiency of genome editing. Researchers worldwide understand the promise of this technology and many have adopted it within their work.
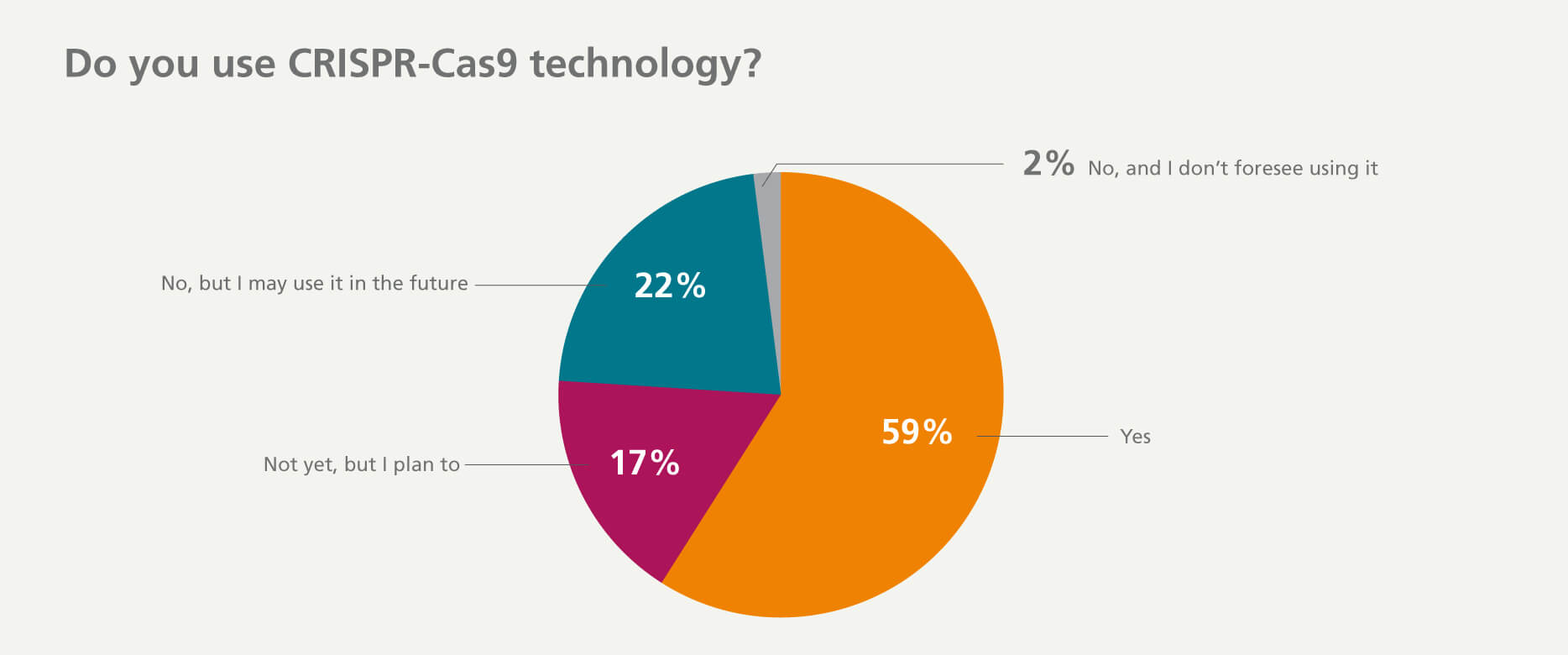
59% of respondents have used CRISPR-Cas9 technology in the last five years and another 17% plan to use it in their upcoming research projects.
An Endless Array of Applications
The ever-expanding CRISPR toolbox has been used in a variety of research areas and experimental systems, including organoids, stem cells, and many primary cell types. The most common research applications among respondents was studying gene function and generating cellular disease models. The variety of applications and wide range of research areas that CRISPR technology were being applied to show the scale of its adoption and impact.
By combining CRISPR and stem cell technologies to introduce or correct pathogenic mutations, researchers are able to study gene function and develop physiologically relevant human disease models. Genome-edited human stem cells have been successfully differentiated in vitro to the endodermal, mesodermal, and ectodermal cell types. CRISPR is also being applied to various organoids, including intestinal and cerebral organoids. Organoid cultures recapitulate the genetic and functional features of in vivo tissue systems, making them a highly informative way to examine pre-existing knockout models to probe the mechanisms of specific cellular processes or disease pathologies. CRISPR technology is also being used in various therapeutic strategies, such as ex vivo editing of human pluripotent stem cells (hPSCs) or primary cell types(e.g. immune cells or hematopoietic stem and progenitor cells (HSPCs) to provide a cell source for clinically relevant cellular therapies.
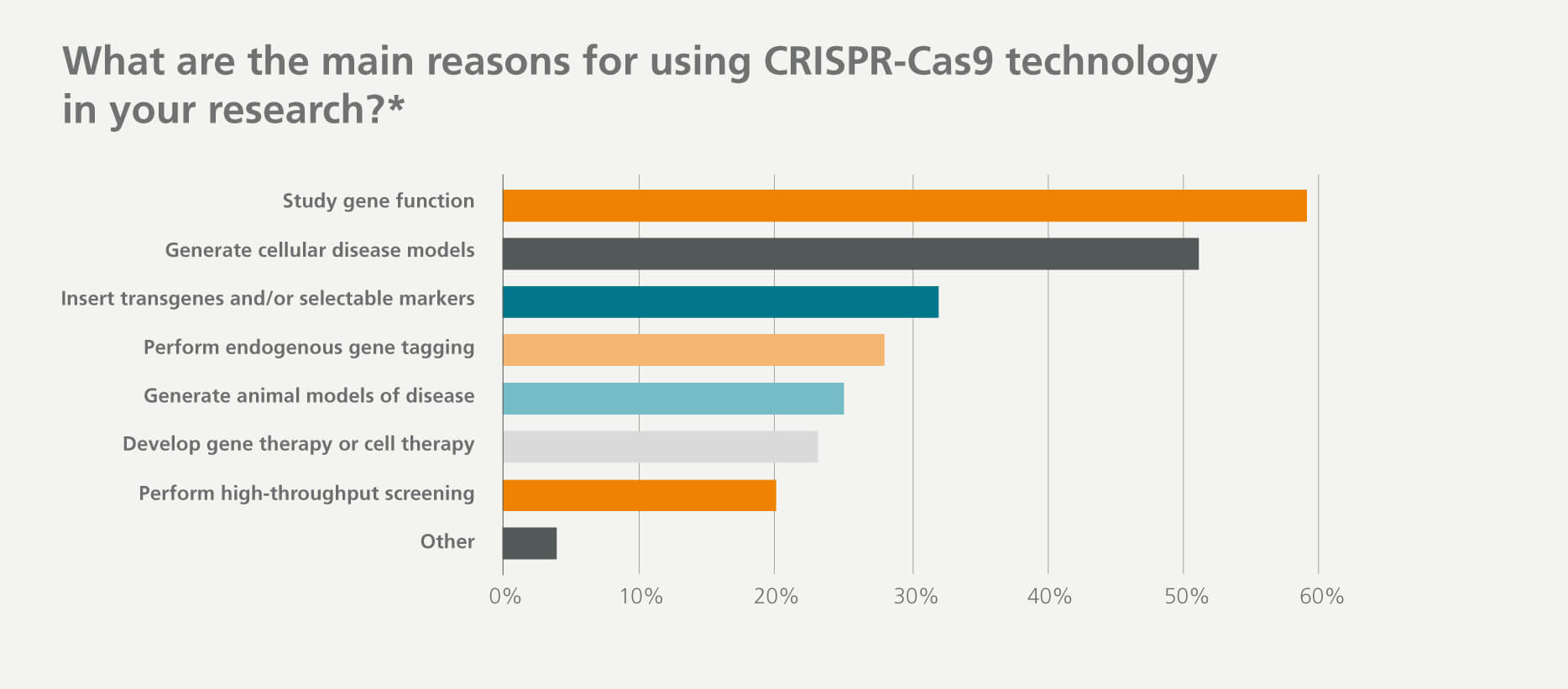
*Respondents were permitted to select multiple answers. Percentages represent the proportion of total respondents who indicated each answer.
~ 1 in 2 Respondents Are Uncomfortable in Applying CRISPR
It is indisputable that CRISPR has already helped to advance our understanding of basic biological mechanisms and to generate advanced cellular and animal disease models. However, significant hurdles will need to be addressed to realize the full potential of this technology. When respondents were asked about how comfortable they feel about applying CRISPR-Cas9 technology in their own lab, 58% of researchers indicated that there are still a few challenges or many challenges in adopting CRISPR-Cas9 technology (see data below). Surprisingly, of the respondents who are currently using CRISPR-Cas9, only 48% find it comfortable to perform genome editing, while the remaining 52% find it somewhat uncomfortable or uncomfortable (data not shown).
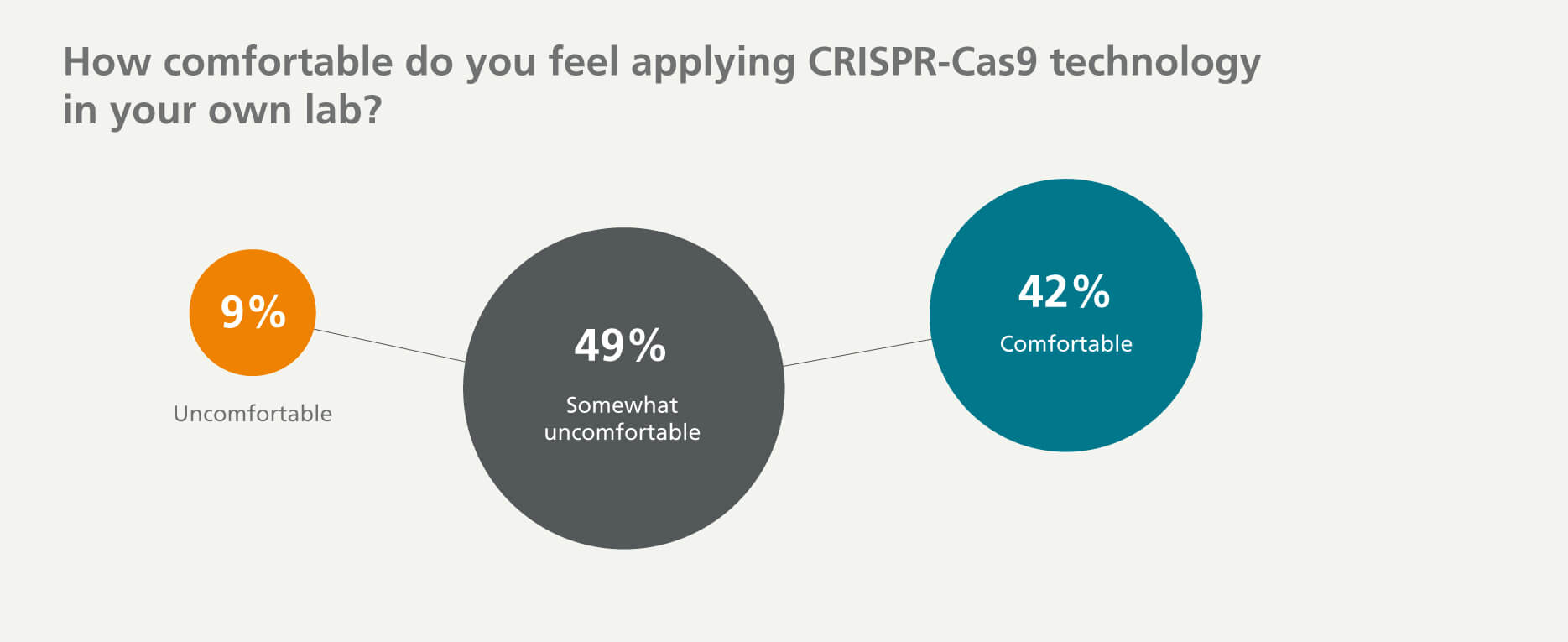
It is an important topic and we need to find more ways to make this technique easier and overcome its challenges.
Survey Respondent
Researchers Prefer to Perform Gene Editing Themselves
Despite the challenges that exist, 54% of researchers indicated that they would perform genome editing in their own lab rather than purchase edited cell lines from a service provider or core facility.
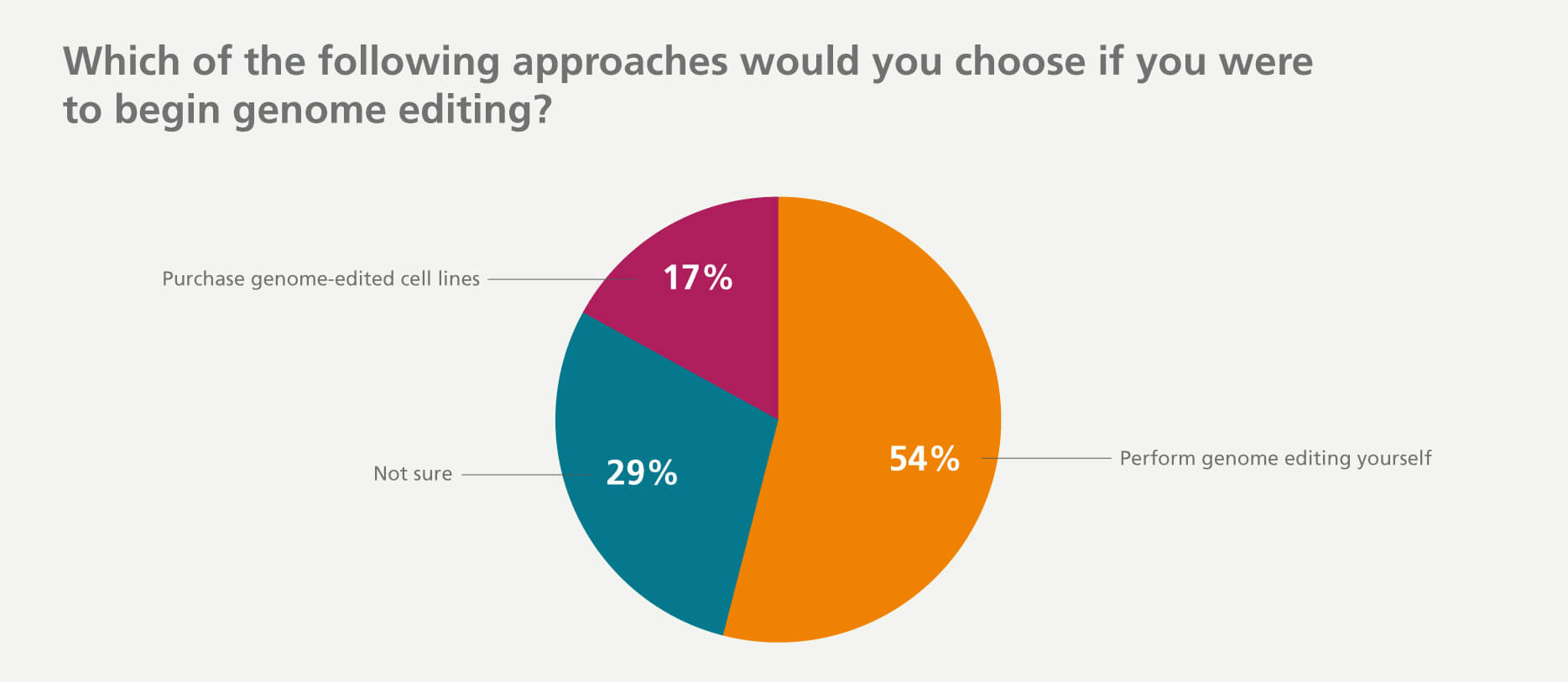
It is a useful tool for many kinds of research projects, and knowing how to perform CRISPR-Cas9 gene editing will aid in evaluating many questions.
Survey Respondent
Facing the Challenges of High-Efficiency CRISPR-Cas9 Gene Editing
Researchers want to perform genome editing in their own lab and understand the benefit of applying CRISPR technology within their research. However, we found that roughly 1 in 2 of all respondents—including 48% of respondents who are currently using CRISPR-Cas9—are uncomfortable applying this technology. To explore challenges and inform solutions, we delved deeper into the key challenges that researchers are facing in performing high-efficiency genome editing. The results of those queries are summarized in the next sections.
The three most prominent concerns brought up in the survey were:
- Achieving optimal genome editing efficiency
- Achieving optimal transfection efficiency
- Maintaining cell viability and function following genome editing
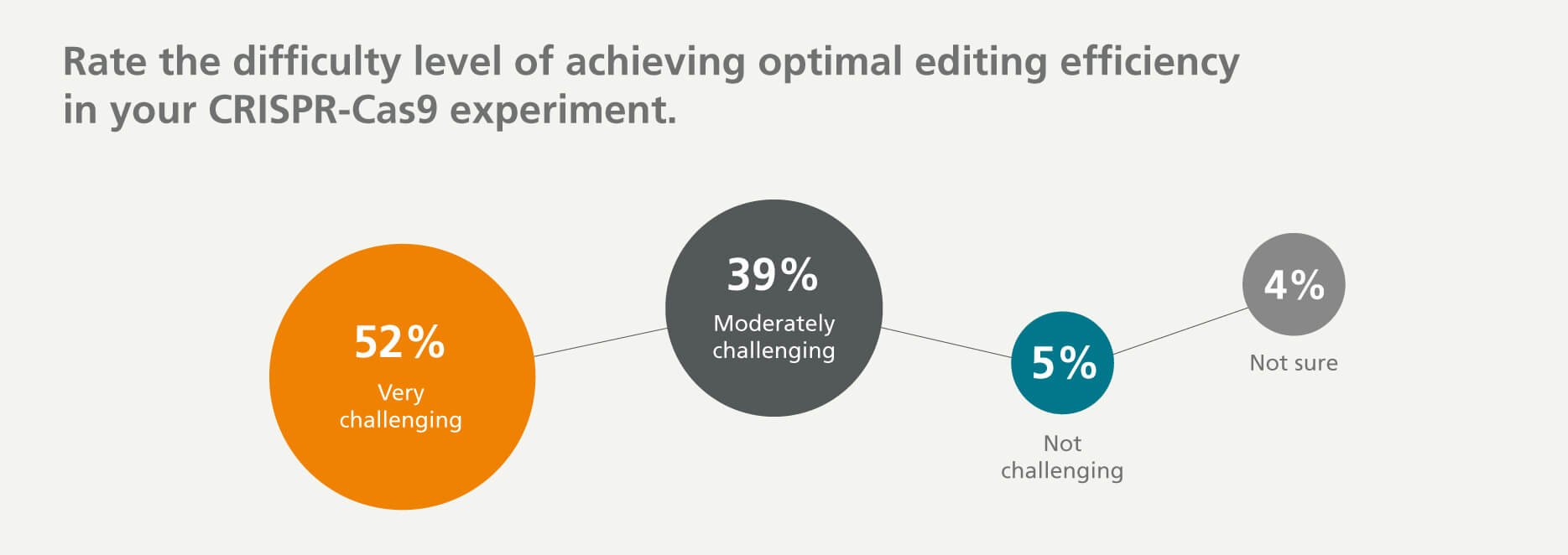
Genome Editing Efficiency is a Key Concern
The goal of genome editing is to generate on-target DNA changes with high efficiency (defined as the percentage of cells carrying the intended mutation in the total population of edited cells). This is impacted by various factors, including gRNA efficiency, cell type-dependent differences in the efficiencies of homology-directed repair (HDR) and non-homologous end joining (NHEJ) repair, cell culture conditions that affect cell growth and viability, and the methods used to deliver and express CRISPR components.
Despite an expanding portfolio of CRISPR tools, achieving high genome editing efficiency remains challenging. More than 50% of respondents indicated that it is very challenging to achieve optimal gene editing efficiency.
Choose the Correct Expression System for CRISPR Components
Researchers can choose from a variety of different CRISPR-Cas expression formats and their choice can greatly impact editing efficiency. The guide RNA (gRNA) and Cas9 proteins can be delivered to the cells in various forms:
- Cas9 and gRNA are encoded on an expression plasmid (DNA-based format) (Figure A).
- Cas9 can be delivered as an mRNA transcript along with an in vitro transcribed (IVT) gRNA (Figure B).
- Purified cas9 protein and custom synthetic gRNA, which form a ribonucleoprotein (RNP) complex, are delivered inside a cell (Figure C). In this format, the gRNA can be composed of a CRISPR RNA (crRNA) and tracrRNA annealed as a crRNA:tracrRNA duplex, or a single guide RNA (sgRNA).
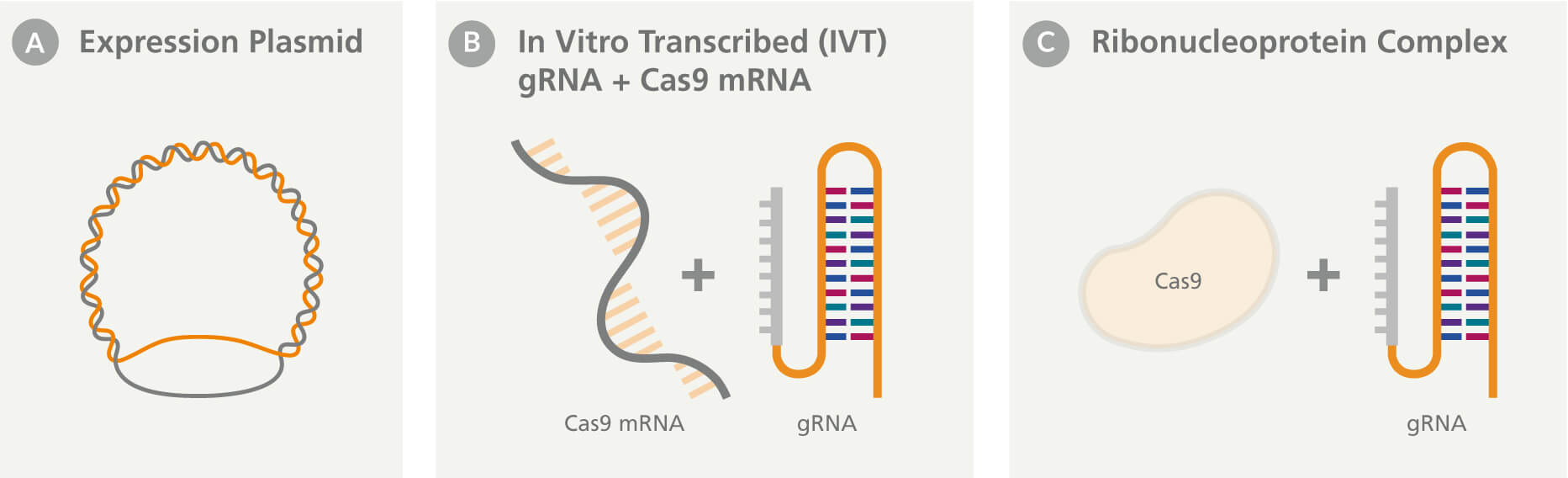
Upon successful delivery, DNA- and RNA-based formats require transcription of the gRNA and Cas9 gene (DNA-based format) and/or translation of the Cas9 protein, which then needs to translocate to the nucleus. Alternatively, the RNP-based system does not require any transcription or translation and nuclear localization sequences (NLS) are added to purified Cas9 protein to enable rapid translocation of the RNP complex to the nucleus upon delivery. Chemical modifications of the gRNA (sgRNA and crRNA:tracrRNA) at various locations can improve stability and increase editing efficiency, for example, 2’-O-methyl and phosphorothioate modifications at the first three 5' and 3' terminal residues.
The RNP format, which has been found to have superior editing efficiency compared to technologies using plasmid or mRNA-based systems, has gained popularity in recent years.1 RNP complexes are immediate acting, exhibit transient expression, and are DNA-free. These features reduce the risk of off-target effects, including Cas9-mediated damage at non-target sites and random integration of plasmid DNA in the genome of target cells. The delivery of nucleic acids is cytotoxic to many primary and stem cell types, which are therefore more amenable to genome editing with RNP-based formats.
For a thorough guide on genome editing experimental design, troubleshooting, and various applications of CRISPR technology, explore the e-book: Genome Editing Applications for Disease Modeling and Cell Therapy, developed in collaboration with Wiley and Current Protocols.
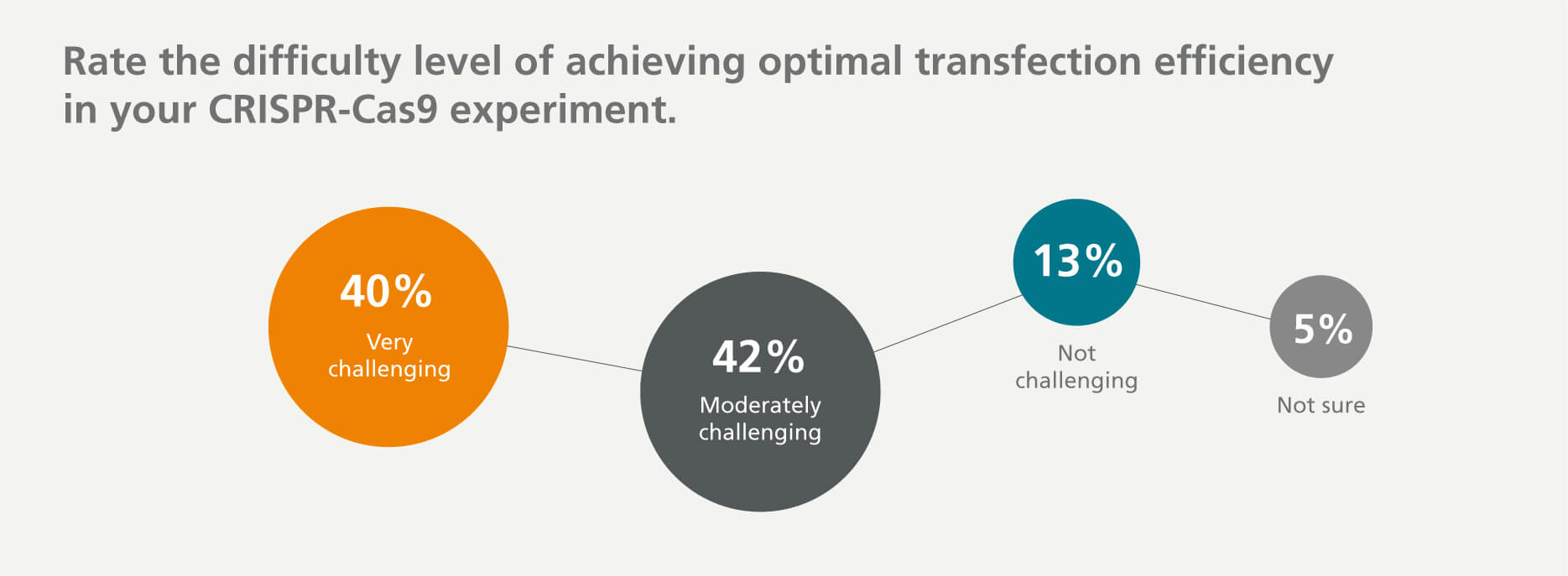
Achieving Optimal Delivery Efficiency of CRISPR Components
Regardless of how the CRISPR components are expressed, one of the greatest challenges is delivering the components across the cell membrane. 40% of respondents agreed and indicated that they find it very challenging to achieve optimal delivery efficiency of CRISPR components.
Chemical transfection and electroporation are the most common methods of CRISPR cargo delivery using non-viral strategies. These delivery methods are relatively inexpensive, straightforward, and effective for genome editing both stem and primary cells. The choice of the delivery system is critical for the cell type of interest as stem cells and many primary cell types such as T cells are difficult to genetically manipulate. For instance, chemical transfection methods can exhibit minimal efficacy in primary immune cell types, often making electroporation a preferred method of delivery. However, electroporation can be harsh on cells, resulting in reduced cell survival and growth post-electroporation. It is therefore critical to optimize delivery conditions for your desired cell type to maximize delivery efficiency while minimizing unwanted side effects such as cell death.
The distinct characteristics of each delivery system make them better suited than the other for certain applications. Electroporation is an effective delivery method for generating in vitro cellular models, however it cannot be used to deliver components in vivo. Viral vector-based approaches to deliver CRISPR components can also be used for both cell culture and in vivo applications. However, viral vectors pose concerns about safety in clinical translation due to the risk of unwanted genetic mutations and immunogenicity.2 In experiments such as oocyte genome editing, where only a small number of cells are to be edited, mRNA encoding Cas9 or RNP complexes are typically preferred since they can be injected directly into the cell of interest and bypass Cas9 and gRNA transcription. This results in faster expression rates and higher genome editing efficiency. For optimal editing efficiency, it is recommended to test the efficiency of CRISPR Cas delivery systems in parallel in target cells or the tissue of interest to determine the optimal method for subsequent experiments.
To learn more about optimized CRISPR-Cas9 genome editing workflows for difficult-to-manipulate cell types, such as T cells and hPSCs, watch this webinar on Genome Editing of Stem and Primary Cells by Dr. Ashley Watson.
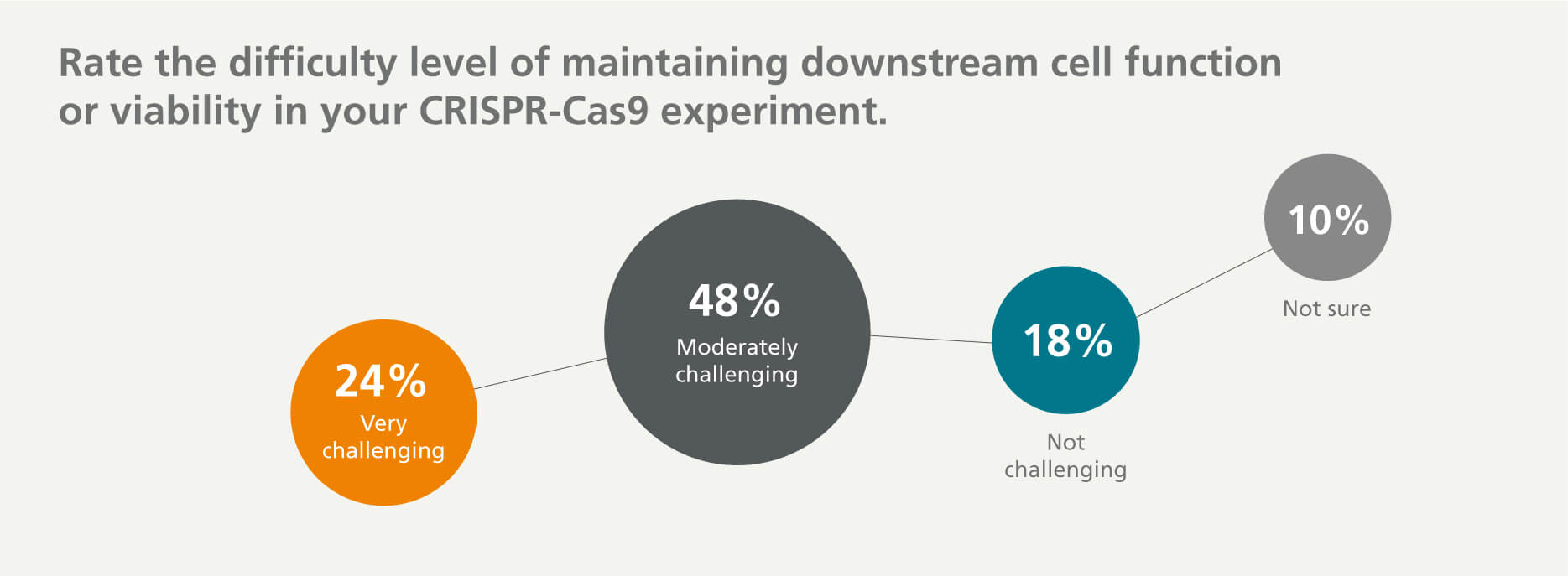
Maintaining Downstream Cell Viability and Functionality
The cellular response to genome editing remains poorly characterized and may negatively impact key features of stem and primary cells such as survival, proliferative capacity, and downstream function. 72% of respondents indicated that it is very challenging or moderately challenging to maintain cell viability or function following CRISPR-Cas9 genome editing.
Relevant and reproducible assays should be used to ensure that genetically manipulated stem and progenitor cells maintain cell identity and viability. Long-term culture of these cell types can result in spontaneous differentiation and loss of potency. Furthermore, genome editing imparts a high degree of selective pressure on cells, especially during post-editing survival.
Studies in hPSCs and HSPCs have observed that activation of the DNA damage response can lead to cell cycle arrest or apoptosis—which constrains editing efficiency—and mutations in proteins required for a correct DNA damage response, such as p53, can be selected for during the genome editing workflow.3-6 It is therefore important to monitor various aspects of cellular health after genome editing and prior to downstream use. Genomic instability is inherent in hPSCs, and it is therefore important to consider the impact of genomic integrity and proactively assess cells after undergoing stress, including from genome editing. This can be done by rapid assessment for common genetic abnormalities or a simple in vitro differentiation assay. In HSPCs, the functional impact of genome editing on lineage commitment can be evaluated using the colony-forming unit (CFU) assay. The CFU assay can be used to assess proliferation and differentiation ability of individual cells within a sample. Furthermore, optimization of pre- and post-editing culture conditions can help minimize experimental variability to support generation of functional and viable edited cells.
Conclusion
As the application of CRISPR technology continues to grow, current and future challenges must be addressed, including achieving optimal editing and delivery efficiency across a wide range of cell types while maintaining cell function and viability. While the field continues to develop strategies to overcome these challenges, it is clear that researchers want to be empowered with the tools and resources to conduct genome editing within their own lab and are inspired by the possibility that CRISPR-Cas9 can help them achieve their next big breakthrough.
Explore validated protocols, webinars, and products to perform high-efficiency genome editing in a variety of cell lines. The protocols include a detailed case study, data, and general tips to overcome challenges associated with genome editing:
Human Pluripotent Stem Cells
Protocol for gene knockout and knock-in in hPSCs using the ArciTect™ CRISPR-Cas9 system, TeSR™ media, and CloneR™.
Intestinal Organoids
Protocol for gene knockout in adult-stem-cell-derived intestinal organoids using the ArciTect™ CRISPR-Cas9 system and IntestiCult™ Organoid Growth Medium (Human).
Hematopoietic Stem Cells
Protocol for gene knockout in CD34+ HSPCs using the ArciTect™ CRISPR-Cas9 system and StemSpan™ Media.
Primary Immune Cells
Protocol for gene knockout in primary human T cells using the ArciTect™ CRISPR-Cas9 system and ImmunoCult™.
The ArciTect™ RNP-based system is a complete CRISPR-Cas9 genome editing platform for editing of various cell lines, including stem cells, primary cell types, and more.
ArciTect™ CRISPR Human Optimization Kit
Optimize your genome editing using this flow-cytometry-based kit validated in stem and primary cell types, including hPSCs, human primary T cells, and CD34+ HSPCs.
ArciTect™ CRISPR Cas9 Genome Editing System
ArciTect™ is a simple and easy-to-use platform to generate functional gene-edited cells in your own lab with validated reagents, cell type-specific protocols, and an integrated Guide RNA Design Tool.
References
- Liang X et al. (2015) Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 208: 44–53. DOI: 10.1016/j.jbiotec.2015.04.024.
- Yin H et al. (2014) Non-viral vectors for gene-based therapy. Nat Rev Genet 15: 541–55. DOI: 10.1038/nrg3763.
- Ihry RJ et al. (2018) p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 24: 939–46. DOI: 10.1038/s41591-018-0050-6.
- Haapaniemi E et al. (2018) CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 24: 927–30. DOI: 10.1038/s41591-018-0049-z.
- Conti A & Di Micco R. (2018) p53 activation: a checkpoint for precision genome editing? Genome Med 10: 66. DOI: 10.1186/s13073-018-0578-6.
- Schiroli G et al. (2019) Precise gene editing preserves hematopoietic stem cell function following transient p53-mediated DNA damage response. Cell Stem Cell 24: 551-65 e8. DOI: 10.1016/j.stem.2019.02.019.